Assignment 3— Solution
Question 1
I read in the data, convert Density to a factor, and fit the two-factor analysis of variance model with Eggs as the response and Season, Density, and their interaction as predictors as in Assignment 1.
quinn <- read.csv( 'ecol 563/quinn1.csv')
quinn$Density <- factor(quinn$Density)
out1 <- lm(Eggs ~ Density*Season, data=quinn)
I next obtain the data frame of means and 95% confidence intervals that we constructed in Assignment 2. The easiest way to obtain the means is to trick R into giving them to us by fitting the following model in which we remove the intercept and include only the interaction term. Alternatively you can use the predict function as was shown in the Assignment 2 solutions.
out1a <- lm(Eggs ~ Density:Season-1, data=quinn)
coef(out1a)
Density6:Seasonspring Density12:Seasonspring Density24:Seasonspring
1.1113333 1.1110000 0.6946667
Density6:Seasonsummer Density12:Seasonsummer Density24:Seasonsummer
3.8866667 2.8866667 2.0000000
fac.vals <- expand.grid(Density=c(6,12,24), Season=c('spring','summer'))
fac.vals
Density Season
1 6 spring
2 12 spring
3 24 spring
4 6 summer
5 12 summer
6 24 summer
fac.vals$Density <- factor(fac.vals$Density)
part1 <- data.frame(fac.vals, ests=coef(out1a), se=summary(out1a)$coefficients[,2], low95=confint(out1a)[,1], up95=confint(out1a)[,2])
part1
Density Season ests se low95 up95
Density6:Seasonspring 6 spring 1.1113333 0.2183934 0.6354950 1.587172
Density12:Seasonspring 12 spring 1.1110000 0.2183934 0.6351616 1.586838
Density24:Seasonspring 24 spring 0.6946667 0.2183934 0.2188283 1.170505
Density6:Seasonsummer 6 summer 3.8866667 0.2183934 3.4108283 4.362505
Density12:Seasonsummer 12 summer 2.8866667 0.2183934 2.4108283 3.362505
Density24:Seasonsummer 24 summer 2.0000000 0.2183934 1.5241616 2.475838
The only new part of this problem is to obtain the difference-adjusted confidence levels so that we can make pairwise comparisons between the means. For that we need the ci.func function that was provided you in lecture 5.
# function to calculate difference-adjusted confidence intervals
ci.func <- function(rowvals, lm.model, glm.vmat) {
nor.func1a <- function(alpha, model, sig) 1-pt(-qt(1-alpha/2, model$df.residual) * sum(sqrt(diag(sig))) / sqrt(c(1,-1) %*% sig %*%c (1,-1)), model$df.residual) - pt(qt(1-alpha/2, model$df.residual) * sum(sqrt(diag(sig))) / sqrt(c(1,-1) %*% sig %*% c(1,-1)), model$df.residual, lower.tail=F)
nor.func2 <- function(a,model,sigma) nor.func1a(a,model,sigma)-.95
n <- length(rowvals)
xvec1b <- numeric(n*(n-1)/2)
vmat <- glm.vmat[rowvals,rowvals]
ind <- 1
for(i in 1:(n-1)) {
for(j in (i+1):n){
sig <- vmat[c(i,j), c(i,j)]
#solve for alpha
xvec1b[ind] <- uniroot(function(x) nor.func2(x, lm.model, sig), c(.001,.999))$root
ind <- ind+1
}}
1-xvec1b
}
To use this function we need the variance covariance matrix of the means. Using the shortcut approach above I can obtain it by applying vcov to my model of cell means.
vmat <- vcov(out1a)
round(vmat,3)
Density6:Seasonspring Density12:Seasonspring Density24:Seasonspring
Density6:Seasonspring 0.048 0.000 0.000
Density12:Seasonspring 0.000 0.048 0.000
Density24:Seasonspring 0.000 0.000 0.048
Density6:Seasonsummer 0.000 0.000 0.000
Density12:Seasonsummer 0.000 0.000 0.000
Density24:Seasonsummer 0.000 0.000 0.000
Density6:Seasonsummer Density12:Seasonsummer Density24:Seasonsummer
Density6:Seasonspring 0.000 0.000 0.000
Density12:Seasonspring 0.000 0.000 0.000
Density24:Seasonspring 0.000 0.000 0.000
Density6:Seasonsummer 0.048 0.000 0.000
Density12:Seasonsummer 0.000 0.048 0.000
Density24:Seasonsummer 0.000 0.000 0.048
If instead we use the original model with the effect estimates then we have to obtain the variance-covariance matrix by matrix multiplication.
cvec <- function(density,season) c(1,density==12, density==24, season=='summer', (density==12)*(season=='summer'), (density==24)*(season=='summer'))
g <- expand.grid(Density=c(6,12,24), Season=c('spring', 'summer'))
cmat <- t(apply(g, 1, function(x) cvec(x[1], x[2])))
cmat
Density Density Season Density Density
[1,] 1 0 0 0 0 0
[2,] 1 1 0 0 0 0
[3,] 1 0 1 0 0 0
[4,] 1 0 0 1 0 0
[5,] 1 1 0 1 1 0
[6,] 1 0 1 1 0 1
vmat1 <- cmat %*% vcov(out1)%*%t(cmat)
round(vmat1,3)
[,1] [,2] [,3] [,4] [,5] [,6]
[1,] 0.048 0.000 0.000 0.000 0.000 0.000
[2,] 0.000 0.048 0.000 0.000 0.000 0.000
[3,] 0.000 0.000 0.048 0.000 0.000 0.000
[4,] 0.000 0.000 0.000 0.048 0.000 0.000
[5,] 0.000 0.000 0.000 0.000 0.048 0.000
[6,] 0.000 0.000 0.000 0.000 0.000 0.048
Using either approach we get the same answer. We need a confidence level of 85% for all the pairwise comparisons.
ci.func(1:6,out1,vmat1)
[1] 0.8506702 0.8506702 0.8506702 0.8506702 0.8506702 0.8506702 0.8506702 0.8506702
[9] 0.8506702 0.8506702 0.8506702 0.8506702 0.8506702 0.8506702 0.8506702
ci.func(1:6,out1a,vmat)
[1] 0.8506702 0.8506702 0.8506702 0.8506702 0.8506702 0.8506702 0.8506702 0.8506702
[9] 0.8506702 0.8506702 0.8506702 0.8506702 0.8506702 0.8506702 0.8506702
I add 85% confidence intervals to data frame of means.
part1$low85 <- part1$ests+qt((1-.85)/2, out1$df.residual) * part1$se
part1$up85 <- part1$ests+qt(1-(1-.85)/2, out1$df.residual) * part1$se
part1
part1
Density Season ests se low95 up95 low85 up85
Density6:Seasonspring 6 spring 1.1113333 0.2183934 0.6354950 1.587172 0.7754538 1.447213
Density12:Seasonspring 12 spring 1.1110000 0.2183934 0.6351616 1.586838 0.7751204 1.446880
Density24:Seasonspring 24 spring 0.6946667 0.2183934 0.2188283 1.170505 0.3587871 1.030546
Density6:Seasonsummer 6 summer 3.8866667 0.2183934 3.4108283 4.362505 3.5507871 4.222546
Density12:Seasonsummer 12 summer 2.8866667 0.2183934 2.4108283 3.362505 2.5507871 3.222546
Density24:Seasonsummer 24 summer 2.0000000 0.2183934 1.5241616 2.475838 1.6641204 2.335880
I now just repeat the graph from Question 3 of Assignment 2. The only change is that I add error bars corresponding to the 85% confidence intervals. The changes are highlighted below.
part1$density.num <- as.numeric(part1$Density)
results.mine <- part1
par(lend=1)
plot(c(.8,3.2), range(results.mine[,c("up95", "low95")]), type='n', xlab="Density", ylab='Mean egg count', axes=F)
axis(1, at=1:3, labels=levels(results.mine$Density))
axis(2)
box()
# Spring
cur.dat <- results.mine[results.mine$Season=='spring',]
myjitter <- 0
arrows(cur.dat$density.num+myjitter, cur.dat$low95, cur.dat$density.num+myjitter, cur.dat$up95, angle=90, code=3, length=.05, col=1)
segments(cur.dat$density.num+myjitter, cur.dat$low85, cur.dat$density.num+myjitter, cur.dat$up85, col='grey70', lwd=3)
lines(cur.dat$density.num+myjitter, cur.dat$ests, col=1)
points(cur.dat$density.num+myjitter, cur.dat$ests, col=1, pch=16)
# Summer
cur.dat <- results.mine[results.mine$Season=='summer',]
myjitter <- 0
arrows(cur.dat$density.num+myjitter, cur.dat$low95, cur.dat$density.num+myjitter, cur.dat$up95, angle=90, code=3, length=.05, col=2)
segments(cur.dat$density.num+myjitter, cur.dat$low85, cur.dat$density.num+myjitter, cur.dat$up85, col='pink2', lwd=3)
lines(cur.dat$density.num+myjitter, cur.dat$ests, col=2)
points(cur.dat$density.num+myjitter, cur.dat$ests, col='white', pch=16, cex=1.1)
points(cur.dat$density.num+myjitter, cur.dat$ests, col=2, pch=1)
legend('topright', c('Spring','Summer'), col=1:2, pch=c(16,1), cex=.8, pt.cex=.9, title=expression(bold('Season')), bty='n')
abline(h=seq(0,4.5,.5), lty=3, col='grey70')
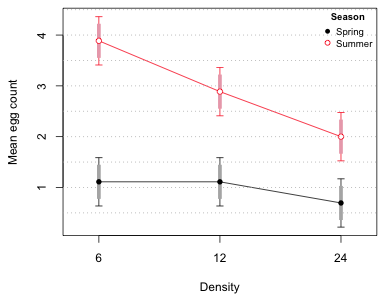
Fig. 1 Interaction plot with difference-adjusted confidence intervals
Based on the display we see that none of the mean egg counts at different densities in Spring are significantly differnet from each other. But in summer the mean egg counts at the three densities are all different from each other. Of course we already knew the spring means and summer means were different because the 95% confidence intervals didn't overlap.
Question 2
pines <- read.delim('ecol 563/pines.txt')
pines[1:8,]
diam species location
1 23 A 1
2 15 A 1
3 26 A 1
4 13 A 1
5 21 A 1
6 28 B 1
7 22 B 1
8 25 B 1
Part a
From the description we should probably treat this as a randomized block design in which location defines the block and species is the treatment. Because we have multiple individuals of the same species at each location we are able to test for a treatment × block interaction.
out1 <- lm(diam~factor(location)*species, data=pines)
anova(out1)
Analysis of Variance Table
Response: diam
Df Sum Sq Mean Sq F value Pr(>F)
factor(location) 3 68.05 22.683 1.2726 0.294392
species 2 284.93 142.467 7.9925 0.001009 **
factor(location):species 6 157.60 26.267 1.4736 0.207343
Residuals 48 855.60 17.825
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
The interaction is not significant so we can fit the standard randomized block design model.
out2 <- lm(diam~factor(location)+species, data=pines)
anova(out2)
Analysis of Variance Table
Response: diam
Df Sum Sq Mean Sq F value Pr(>F)
factor(location) 3 68.05 22.683 1.2089 0.315317
species 2 284.93 142.467 7.5930 0.001242 **
Residuals 54 1013.20 18.763
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
We find the species effect is significant. To see which species differ we can examine the summary table.
round(summary(out2)$coefficients,3)
Estimate Std. Error t value Pr(>|t|)
(Intercept) 19.167 1.370 13.993 0.000
factor(location)2 1.400 1.582 0.885 0.380
factor(location)3 2.000 1.582 1.264 0.211
factor(location)4 -0.667 1.582 -0.421 0.675
speciesB 3.200 1.370 2.336 0.023
speciesC -2.100 1.370 -1.533 0.131
So, species A and B are significantly different but species A and C are not. Because the mean diameter of species B exceeds A whereas the mean diameter of species C is less than A, it's pretty clear that species B is also different from species C. To test this formally we can refit the model choosing a different species as the reference group. I choose species B as the reference group
out2a <- lm(diam~factor(location) + factor(species, levels=c('B','A','C')), data=pines)
round(summary(out2a)$coefficients,3)
Estimate Std. Error t value Pr(>|t|)
(Intercept) 22.367 1.370 16.329 0.000
factor(location)2 1.400 1.582 0.885 0.380
factor(location)3 2.000 1.582 1.264 0.211
factor(location)4 -0.667 1.582 -0.421 0.675
factor(species, levels = c("B", "A", "C"))A -3.200 1.370 -2.336 0.023
factor(species, levels = c("B", "A", "C"))C -5.300 1.370 -3.869 0.000
Now we see that species C is significantly different from species B also.
Note: In a randomized block design one generally does not test for a block effect. In most situations the reported F-statistic for the block effect does not truly have an F-distribution so the reported p-value is incorrect. Even when the F-distribution of the statistic holds we are forced to make the additional assumption that the individuals were randomly assigned to blocks, which is usually not the case. The larger point is that blocks are a part of the design structure of the experiment; they are not part of the treatment structure. Thus blocks should be retained regardless of what the significance tests suggest. For further discussion on this point see lecture 7.
Part b
In the fixed effects version of the randomized block design while we can estimate species differences but we can only estimate the mean diameters of individual species for specific blocks. To get around this we can treat block as a random effect in which case the regression model is formulated for the average individual of each species. In the mixed effects model the block effects (random effects) represent deviations from that average.
Method 1: Using the nlme package plus hand calculations
We can calculate the confidence intervals for the means by hand as follows. I first fit the model using lme.
out2.lme <- lme(diam~species, random=~1|location, data=pines)
The degrees of freedom we need for the t-distribution are contained in the fixDF component of the model in a term labeled X.
names(out2.lme)
[1] "modelStruct" "dims" "contrasts" "coefficients" "varFix" "sigma"
[7] "apVar" "logLik" "numIter" "groups" "call" "terms"
[13] "method" "fitted" "residuals" "fixDF" "na.action" "data"
out2.lme$fixDF
$X
(Intercept) speciesB speciesC
54 54 54
$terms
(Intercept) species
54 54
attr(,"assign")
attr(,"assign")$`(Intercept)`
[1] 1
attr(,"assign")$species
[1] 2 3
attr(,"varFixFact")
[,1] [,2] [,3]
[1,] 0.6148623 0.0000000 0.00000
[2,] 0.3954213 -1.1862640 0.00000
[3,] 0.6848898 -0.6848898 -1.36978
Next I construct a vector that can be used to construct the terms of the regression model.
fixef(out2.lme)
(Intercept) speciesB speciesC
19.85 3.20 -2.10
To get the contrast matrix I sapply this function on a vector of species names. I then use the contrast matrix to obtain the means and standard errors of the means in the usual fashion.
cmat <- t(sapply(c('A','B','C'), cvec))
est <- cmat %*% fixef(out2.lme)
se <- sqrt(diag(cmat %*% vcov(out2.lme) %*% t(cmat)))
low95 <- est + qt(.025, out2.lme$fixDF$X)*se
up95 <- est + qt(.975, out2.lme$fixDF$X)*se
data.frame(est, low95, up95)
est low95 up95
A 19.85 17.84163 21.85837
B 23.05 21.04163 25.05837
C 17.75 15.74163 19.75837
Method 2: Using the nlme package and the intervals function
If we fit the random effects model without the intercept term, lme returns the mean diameter for each species. We can then obtain 95% confidence intervals for the treatment means by using the intervals function.
out2a.lme <- lme(diam~species-1, random=~1|location, data=pines)
intervals(out2a.lme, which="fixed")
Approximate 95% confidence intervals
Fixed effects:
lower est. upper
speciesA 17.84163 19.85 21.85837
speciesB 21.04163 23.05 25.05837
speciesC 15.74163 17.75 19.75837
attr(,"label")
[1] "Fixed effects:"
Method 3: Using the lmer package and obtaining means via MCMC sampling
Using the lme4 package we can generate Bayesian samples from the posterior distributions of the parameters in the model. These can then be used to construct the means and their confidence (credible) intervals.
detach(package:nlme)
library(lme4)
out2.lmer <- lmer(diam~species+ (1|location), data=pines)
Obtain 10,000 samples from the posterior distribution of each parameter.
out2.mc <- mcmcsamp(out2.lmer, n=10000)
mc.dat <- as.data.frame(out2.mc)
names(mc.dat)
[1] "(Intercept)" "speciesB" "speciesC" "ST1" "sigma"
To obtain the posterior distributions of the means we just have to construct the means from their effect estimates. The mean of species A is the intercept in the model which is column 1, the mean of species B is the intercept plus the species B effect (column 1 plus column 2), and the mean of species C is the intercept plus the species C effect (column 1 plus column 3).
new.dat <- data.frame(mc.dat[,1], mc.dat[,1] + mc.dat[,2], mc.dat[,1] + mc.dat[,3])
Finally I obtain the .025 and .975 quantiles of each column of new.dat to obtain the 95% percentile credible intervals for the means. For a point estimate of the mean I can take either the mean or the median of the individual posterior distributions.
apply(new.dat, 2, function(x) quantile(x,c(.025,.975)))
mc.dat...1. mc.dat...1....mc.dat...2. mc.dat...1....mc.dat...3.
2.5% 17.34762 20.51541 15.24718
97.5% 22.35942 25.57697 20.28810
out.95 <- apply(new.dat, 2, function(x) quantile(x, c(.025,.975)))
out.mean <- apply(new.dat, 2, mean)
out.median <- apply(new.dat, 2, median)
results <- data.frame(species=c('A','B','C'), mean=out.mean, out.95, median=out.median)
rownames(results) <- NULL
results
species mean X2.5. X97.5. median
1 A 19.82780 17.34762 22.35942 19.81285
2 B 23.06143 20.51541 25.57697 23.05437
3 C 17.75208 15.24718 20.28810 17.74412
Course Home Page


