Analysis of covariance is just a regression model in which the main predictor(s) of interest is (are) categorical, but a potentially confounding continuous variable was also measured. Thus analysis of covariance is analysis of variance in which there is one or more additional continuous covariates. The terminology dates from a time when it was not generally appreciated that regression is the common theme that links analysis of variance and regression models together.
There are two basic uses for analysis of covariance and related models.
The example we'll consider appears in Crawley (2002), p. 287, (also Crawley 2007) where it is described as follows.
"The next worked example concerns an experiment on the impact of grazing on the seed production of a biennial plant (Ipomopsis). Forty plants were allocated to treatments, grazed and ungrazed, and the grazed plants were exposed to rabbits during the first two weeks of stem elongation. They were then protected from subsequent grazing by the erection of a fence and allowed to regrow. Because initial plant size was thought likely to influence fruit production, the diameter of the top of the rootstock was measured before each plant was potted up. At the end of the growing season, the fruit production (dry weight, mg) was recorded on each of the 40 plants, and this forms the response variable for the analysis."
There are three recorded variables.
The goal is to determine the effect that grazing on plants when they're young has on their fruit production when they're adults.
To test for a grazing effect on fruit production we carry out an analysis of variance.
Response: Fruit
Df Sum Sq Mean Sq F value Pr(>F)
Grazing 1 2910.4 2910.44 5.3086 0.02678 *
Residuals 38 20833.4 548.25
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
So there's a significant effect due to grazing. When we examine the estimated effect we see that it's in a surprising direction. Grazed plants have higher fruit production than ungrazed plants.
Residuals:
Min 1Q Median 3Q Max
-52.991 -18.028 2.915 14.049 48.109
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 67.941 5.236 12.976 1.54e-15 ***
GrazingUngrazed -17.060 7.404 -2.304 0.0268 *
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 23.41 on 38 degrees of freedom
Multiple R-squared: 0.1226, Adjusted R-squared: 0.09949
F-statistic: 5.309 on 1 and 38 DF, p-value: 0.02678
It's worth noting that analysis of variance when a categorical predictor has two levels is identical to carrying out a two sample t-test. We can carry out a t-test in R with the t.test function.
data: Fruit by Grazing
t = 2.304, df = 37.306, p-value = 0.02689
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
2.061464 32.058536
sample estimates:
mean in group Grazed mean in group Ungrazed
67.9405 50.8805
data: Fruit by Grazing
t = 2.304, df = 38, p-value = 0.02678
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
2.070631 32.049369
sample estimates:
mean in group Grazed mean in group Ungrazed
67.9405 50.8805
The ANOVA result is identical to the pooled-variances t-test obtained by specifying var.equal=T. The separate variances t-test is equivalent to the variance heterogeneity ANOVA model like the one we fit using gls with a weights argument in lecture 3.
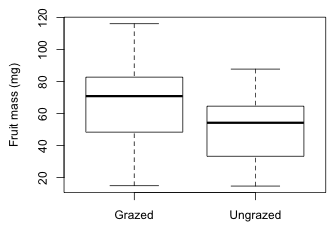
If we plot the distribution of fruit mass in the two groups we see that the distributions are consistent with the analytical results. Grazed plants have a higher average fruit production than do ungrazed plants.

Fig. 1 The effect of grazing on fruit mass
So far we've been ignoring the covariate root size that was also measured as part of the experiment. In Fig. 2 I plot fruit mass versus root size color coding the points according to their grazing status. I use the numeric values of the Grazing factor, 1 and 2, as the argument to col= to specify the color codes 1 and 2. I do the same to get different plotting symbols but this time using 1 and 2 to select either the first or the second element of the vector mypch that specifies open and filled circles.
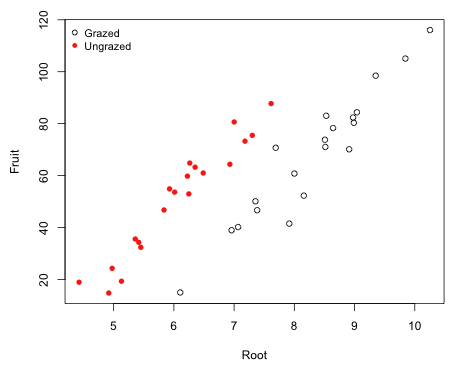
Fig. 2 The effect of grazing on fruit mass while controlling for root size
Observe that the root size distributions are different in the two treatment groups. The plants in the grazed group tend to have larger root sizes that those in the ungrazed group. Also notice that there is a strong linear relationship between fruit production and root size. Thus it seems possible that by not accounting for root size in our analysis that the estimation of a treatment effect has been compromised.
In the biological literature a method that is often used to adjust for a potentially confounding variable such as root size is to create a new response variable that is the ratio of the original response variable and the confounder. Using this approach here we would analyze the ratio of fruit production to root size rather than fruit production. If we do so we find that the fruit production to root size ratio is not significantly different between the two grazing groups.
Response: Fruit/Root
Df Sum Sq Mean Sq F value Pr(>F)
Grazing 1 0.177 0.1773 0.0306 0.862
Residuals 38 219.994 5.7893
Residuals:
Min 1Q Median 3Q Max
-5.4980 -1.6605 0.7814 1.5884 3.4426
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 7.9464 0.5380 14.770 <2e-16 ***
GrazingUngrazed 0.1332 0.7609 0.175 0.862
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 2.406 on 38 degrees of freedom
Multiple R-squared: 0.0008054, Adjusted R-squared: -0.02549
F-statistic: 0.03063 on 1 and 38 DF, p-value: 0.862
Notice that although the point estimate of the grazing effect on the ratio is not significant, it is positive suggesting that ungrazed plots have a larger fruit to root ratio than do grazed plots. This is the reverse of the direction we found when not accounting for root size.
In addition to creating a variable that is harder to interpret than the original variable, the use of a ratio response makes some unwarranted assumptions about the relationship of fruit production to root size. We'll examine these assumptions below after we consider a better way to adjust for root size—analysis of covariance.
Notice that in Fig. 2 the two treatment groups are completely segregated each lying within a distinct band. At any root size where the treatment groups overlap, the fruit production is seen to be higher for the ungrazed group and than for the grazed group. This suggests that there is a treatment effect, although not as simple as an ordinary mean difference between the groups. Given that a linear relationship between fruit mass and root size appears to be appropriate for both treatment groups there are three possible models to consider here.
(a)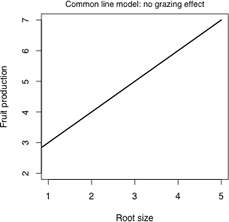 |
(b)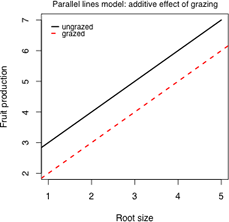 |
(c)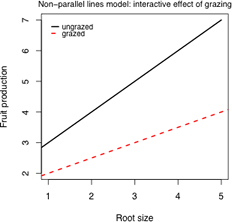 |
| Fig. 3 Three possible regression models for the effect of grazing on fruit production | ||
Let
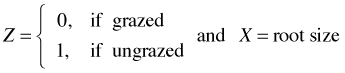
The three models can be expressed as follows.
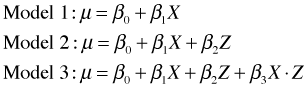
Regression models with categorical variables can also be written as multiple linear equations where there is a separate equation for each category of the categorical variable. A regression model with a single dummy variable yields two different equations, one for each value of the dummy variable. To see the connection between the three models described above and Fig. 3, replace the dummy variable Z by its values 0 and 1.
Model 2: Parallel lines
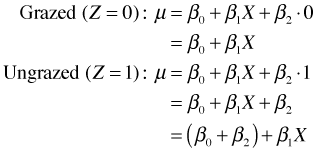
We have the equations of two lines that have the same slope, β1, but different intercepts. The parameter β2 measures how much the intercepts differ. It also corresponds to the vertical distance between the parallel lines in Fig. 3b.
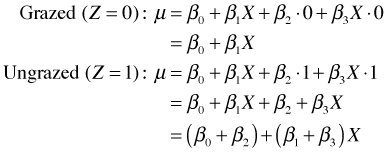
Here we have the equations of two lines with different intercepts and different slopes. The parameter β2 measures how much the intercepts differ. The parameter β3 measures how much the slopes differ.
I fit a sequence of models starting with the single line model, then the additive model, and finally the full interaction model. The additive model is the analysis of covariance model in this example. When we examine model 1 in which root is the only predictor, we see that there is a significant positive effect due to root size.
Next we turn to model 2, the parallel lines model.
Response: Fruit
Df Sum Sq Mean Sq F value Pr(>F)
Root 1 16795.0 16795.0 368.91 < 2.2e-16 ***
Grazing 1 5264.4 5264.4 115.63 6.107e-13 ***
Residuals 37 1684.5 45.5
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
From the output we see that having controlled for root size, there is a significant grazing effect on fruit production. To see the nature of that effect we can examine the summary table of the model.
Residuals:
Min 1Q Median 3Q Max
-17.1920 -2.8224 0.3223 3.9144 17.3290
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -127.829 9.664 -13.23 1.35e-15 ***
Root 23.560 1.149 20.51 < 2e-16 ***
GrazingUngrazed 36.103 3.357 10.75 6.11e-13 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 6.747 on 37 degrees of freedom
Multiple R-squared: 0.9291, Adjusted R-squared: 0.9252
F-statistic: 242.3 on 2 and 37 DF, p-value: < 2.2e-16
According to model 2 the fruits of ungrazed plants are on average 36.1 mg heavier than the fruits of grazed plants. Finally I consider model 3, the non-parallel lines model.
Response: Fruit
Df Sum Sq Mean Sq F value Pr(>F)
Root 1 16795.0 16795.0 359.9681 < 2.2e-16 ***
Grazing 1 5264.4 5264.4 112.8316 1.209e-12 ***
Root:Grazing 1 4.8 4.8 0.1031 0.75
Residuals 36 1679.6 46.7
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
The interaction is not significant thus we can stick with the parallel lines model. Because the interaction is not significant, the linear relationship between fruit mass and root size is the same in both grazing groups. I re-examine the coefficients of the parallel lines model.
The parallel lines model is the classic analysis of covariance model. In this model the grazing effect term adds to the intercept, so the intercept of the fruit-root relationship is 36.1 units larger for ungrazed plants than it is for grazed plants. Because the lines are parallel, 36.1 is also the vertical distance between the lines at any root size. So we can say that ungrazed plants yield fruit that is on average 36.1 mg heavier than the the fruit of grazed plants after controlling for the size of the plant (based on the size of their root). This result is the exact opposite of the one we obtained when we did not control for the confounding effects of root size. The utility of the analysis of covariance model is that it allows us to statistically control for systematic differences between groups so that we can validly compare the means of the groups.
I graph the raw data and superimpose the best model, the parallel lines model (model 2). A convenient way to superimpose functions on already plotted data is with the curve function. The first argument of curve should be a function of the variable x. To force the curve function to add the graph of the function to the currently active graphics window we need to include the add=T argument.
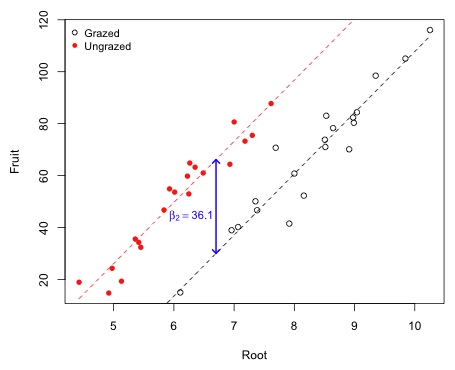
Fig. 4 Analysis of covariance model showing the treatment effect β2
The estimate of the grazing effect on fruit mass corresponds to the vertical distance between the parallel lines in Fig. 4. Generally speaking when graphing regression lines we should not extrapolate beyond the range of the data. I redraw Fig. 4 and truncate the regression lines accordingly using the minimum and maximum values of root size in each grazing group. The curve function has from= and to= arguments that allow you to specify x-axis limits in drawing the function.
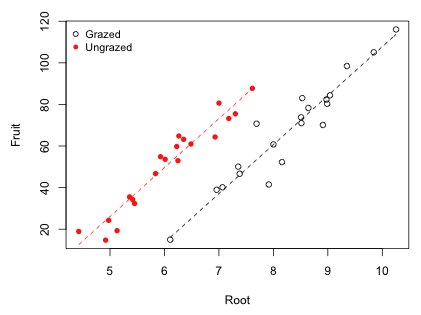
Fig. 5 Analysis of covariance model with regression lines truncated to the range of data
Having adjusted for root size by using the analysis of covariance model let's revisit the regression model that used the ratio of fruit mass to root size as the response variable and see what's wrong with it. If Y is fruit mass, X is root size, and Z is a dummy variable indicating grazing status, then the regression model we fit for the ratio of fruit mass to root size is the following.
|
|
(1) |
|---|
where ![]() . I've used different symbols for the regression coefficients so as not to confuse them with the analysis of covariance model above. Multiply both sides of the equation by X, the root size variable.
. I've used different symbols for the regression coefficients so as not to confuse them with the analysis of covariance model above. Multiply both sides of the equation by X, the root size variable.
|
|
(2) |
|---|
where ε* is just my notation for the new error term. (Note: Because ε is multiplied by X this model assumes that the standard deviation of the residuals is not constant but varies with the magnitude of X. So, we have a heteroscedastic error model.) Compare this last equation with model 3, the non-parallel lines model, described above.
|
|
(3) |
|---|
There are two obvious differences between the models shown in eqns (2) and (3).
Fig. 6 shows the analysis of covariance model of Fig. 4 and superimposes the ratio model of eqn (2) using the coefficients estimated from eqn (1). The badness of the ratio model is readily apparent.
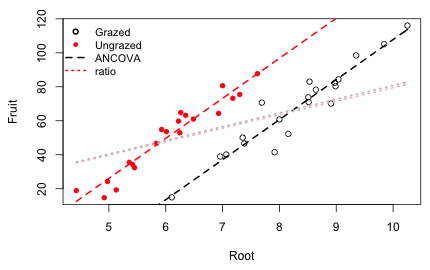
Fig. 6 Analysis of covariance model compared with ratio regression model
Fig. 6 is a bit unfair because fitting the ratio model in eqn (1) using least squares does not yield the same parameter estimates as fitting eqn (2) using least squares (although they are close). Basically the difference is that the mean of a ratio is not equal to the ratio of the means. Still dividing a response variable by a covariate as an attempt to "standardize" it is generally a bad idea. Ratio variables cause problems in ordinary regression because their distribution is typically non-normal. Even worse the use of ratios has the potential of inducing spurious correlations between the variables of interest. Recognition of the problems posed by ratio variables dates back to the beginning of statistics itself (Pearson 1897). Over the years researchers in applied disciplines have repeatedly "rediscovered" the ratio problem. Some recent criticisms of using ratio variables in regression instead of analysis of covariance, organized by discipline, include the following.
Here are two excerpts of the criticisms found in these papers. From Beaupre and Dunham (1995), p. 880:
(The) dangers (of using ratios) include: (1) identification of spurious relationships which may lead to erroneous biological interpretation, (2) false identification of effects as significant, (3) failure to identify significant effects and (4) potentially large errors in ... estimation.
… We believe that our reanalysis suggests that these problems are far worse than most appreciate. We urge physiologists to abandon analyses of ratio-based indices in favour of ANCOVA.
From Packard and Boardman (1999), p. 1:
Ecological physiologists commonly divide individual values for variables of interest by corresponding measures for body size to adjust (or scale) data that vary in magnitude or intensity with body size of the animals being studied. These ratios are formed in an attempt to increase the precision of data gathered in planned experiments or to remove the confounding effects of body size from descriptive studies...
This procedure for adjusting data is based on the implicit, albeit critical, assumption that the variable of interest varies isometrically with body size. Isometry occurs whenever a plot of a physiological variable (y) against a measure of body size (x) yields a straight line that passes through the origin of a graph with linear coordinates. Alternatively allometry obtains whenever such a plot yields either a curved line or a straight line that intersects the Y-axis at some point other than zero. Most physiological variables change allometrically with body size (Smith 1984), so the assumption of isometry that is fundamental to the use of ratios for scaling data seldom is satisfied ... statistical analyses of ratios lead to conclusions that are inconsistent with impressions gained from visual examinations of data displayed in bivariate plots. In comparison, analyses of covariance lead to conclusions that agree with impressions gained from these same plots. We therefore recommend that ecological physiologists discontinue using ratios to scale data and that they use the ANCOVA instead.
The opinion of all of these authors is that ratios should almost never be used for statistical control purposes. Their most important objection is that ratios can correctly adjust for the effect of the covariate only when the the response and the covariate are collinear with an intercept of zero. If the intercept is not zero or the relationship is nonlinear (or linear only over a short range), regression with a ratio response can yield spurious results. While the use of ratios is not always inappropriate, if the ratio itself is not of primary interest but is being used only to adjust the value of the response, then analysis of covariance is a far better approach.
Forrester and Steele (2004) studied the effect of competition and resource availability on the mortality of gobies. Resource availability for gobies was defined to be the density of available refuges that allow gobies to hide from their potential predators. It is expected that mortality will increase as population density increases because of intraspecific competition for refuges. Because each refuge is only used by a single goby, the effect of increasing population density should be lower when the availability of refuges is high. The experiment was conducted in the natural habitat of gobies where refuge density was classified into three categories: low, medium, and high. Six replicates of each of these refuge density categories were obtained and for each replicate a different goby density was maintained. Using a detailed census of the resident fish population, mortality was recorded as percent survival. Because percentages are bounded on the interval 0 to 100 treating them as normally distributed variables can lead to nonsensical results. Forrester and Steele (2004) chose to ignore these complications and for today's lecture we will too.
Unlike the Ipomopsis example considered previously where the focus was to compare means across groups and regression was used only to account for a confounding variable, here the focus is on the regression equation itself. The question of interest is whether the relationship between goby mortality and goby density is different for different categories of refuge density. In the Ipomopsis example we had to first rule out the presence of an interaction between the continuous predictor "root" and the categorical treatment variable "grazing" in order to estimate a treatment effect. In the goby experiment the interaction between the continuous predictor goby density and the categorical predictor refuge density was the focus of the analysis. If a significant interaction is present it means that the relationship between goby mortality and goby density is different for different refuge densities.
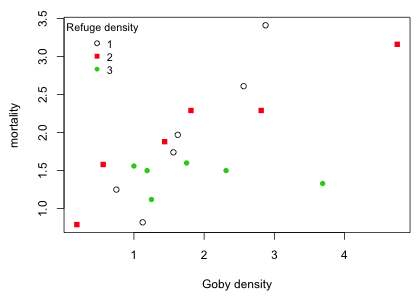
Fig. 7 Effect of goby density and refuge density on goby mortality
The graph would seem to indicate that the goby mortality-goby density relationship varies with refuge density. I fit a model that includes an interaction between refuge density and goby density.
The interaction is statistically significant. The regression equation that was fit is the following. If x = goby density,
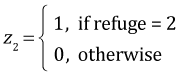 , and
, and 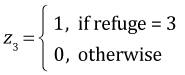
then the predicted mean is
![]()
This reduces to three regression models, one for each refuge type.
| Table 1 Regression model with a continuous predictor and a categorical predictor with three levels |
| refuge | z1 | z2 | |
| 1 | 0 | 0 | 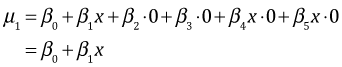 |
| 2 | 1 | 0 | 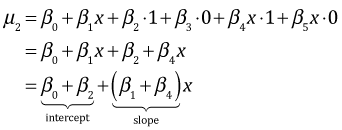 |
| 3 | 0 | 1 | 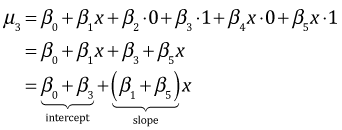 |
Next we examine the summary table of the model.
Residuals:
Min 1Q Median 3Q Max
-0.47439 -0.10553 0.06187 0.15585 0.35897
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.0843 0.3009 0.280 0.784129
density 1.0756 0.1581 6.805 1.89e-05 ***
factor(refuge)2 1.0493 0.3564 2.944 0.012288 *
factor(refuge)3 1.4016 0.4021 3.486 0.004500 **
density:factor(refuge)2 -0.6269 0.1762 -3.558 0.003938 **
density:factor(refuge)3 -1.1029 0.2035 -5.419 0.000155 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.2901 on 12 degrees of freedom
Multiple R-squared: 0.8867, Adjusted R-squared: 0.8395
F-statistic: 18.78 on 5 and 12 DF, p-value: 2.664e-05
From the summary table we see that β4 ≠ 0 (p = .004) so the slopes for refuge =1 and refuge = 2 differ. Because the point estimate of β4 is negative, the slope for refuge 2 is less than the slope for refuge = 1. Also from the summary table we see that β5 ≠ 0 (p = .0002) so the slopes for refuge =1 and refuge = 3 differ. Because the point estimate of β5 is negative, the slope for refuge = 3 is less than the slope for refuge = 1.
To compare the slopes for refuge = 2 and refuge =3 we can refit the model choosing a different reference level for refuge. I make refuge = 3 the reference level in the run below.
Residuals:
Min 1Q Median 3Q Max
-0.47439 -0.10553 0.06187 0.15585 0.35897
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.48587 0.26674 5.571 0.000122 ***
density -0.02728 0.12818 -0.213 0.835038
factor(refuge, levels = 3:1)2 -0.35231 0.32811 -1.074 0.304049
factor(refuge, levels = 3:1)1 -1.40157 0.40211 -3.486 0.004500 **
density:factor(refuge, levels = 3:1)2 0.47603 0.14996 3.174 0.008003 **
density:factor(refuge, levels = 3:1)1 1.10292 0.20351 5.419 0.000155 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.2901 on 12 degrees of freedom
Multiple R-squared: 0.8867, Adjusted R-squared: 0.8395
F-statistic: 18.78 on 5 and 12 DF, p-value: 2.664e-05
With refuge = 3 as the reference level, a test of H0: β4 = 0 is a test of whether the slopes for refuge = 2 and refuge = 3 are the same. From the output we conclude β4 ≠ 0 (p = 0.008). The point estimate is positive so we conclude that the slope for refuge = 2 exceeds the slope for refuge = 1.
To graph the model we can use the abline function applied to lm models fit separately for different values of refuge. The abline extracts the intercept and slope form the lm model object and draws the regression line.
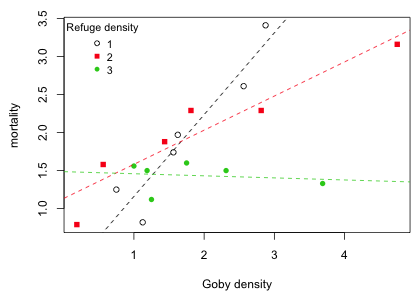
Fig. 8 Regression of mortality on goby density separately by range density
Alternatively we can produce Fig. 8 by extracting the intercepts and slopes ourselves from the full interaction model and then providing them to abline to plot the lines.
Unfortunately abline does not have an option that permits restricting the regression line to the range of the data. For that we could use the curve function like we did in Fig. 5 for the Ipomopsis example. Alternatively we can use the predict function with the newdata argument to obtain the coordinates of the endpoints of the regression line. The range function can be used to calculate the minimum and maximum values of goby density for a given refuge category and the predict function used to obtain the model predictions at those values. I assemble all of this as a function whose sole argument is the refuge density category.
Using the function I add the regression lines to a scatter plot of the data.
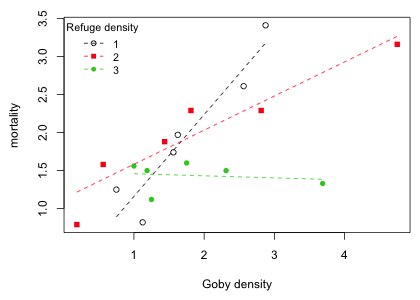 |
| Fig. 9 Regression of goby mortality on goby density separately by refuge density in which regression lines are restricted to the range of the data |
If we attempt to use analysis of covariance to control for a continuous covariate and it turns out that there is a significant interaction between the treatment and the covariate then there is no unique treatment effect. The magnitude (and perhaps the direction) of the treatment effect varies with the value of the continuous covariate. So, in this case we might just summarize the results by reporting the different slopes of the regression lines for the different treatments. Still, we might be interested in knowing if there is a range of covariate values for which a significant treatment effect did occur and perhaps obtain a ranking of the treatments over that range. For example, in Fig. 9 it appears that on the left side of the diagram refuge density doesn't matter, but on the right side of Fig. 9 the percent mortality for different values of refuge can be ranked as follows: mortality(refuge = 1) > mortality(refuge = 2) > mortality(refuge = 3). Can we quantify this result and perhaps summarize it graphically?
Using the trick that was discussed previously, I begin by reparameterizing the model so that the parameter estimates we obtain are estimates of the individual slopes and intercepts.
Residuals:
Min 1Q Median 3Q Max
-0.47439 -0.10553 0.06187 0.15585 0.35897
Coefficients:
Estimate Std. Error t value Pr(>|t|)
factor(refuge)1 0.08430 0.30090 0.280 0.784129
factor(refuge)2 1.13355 0.19108 5.932 6.90e-05 ***
factor(refuge)3 1.48587 0.26674 5.571 0.000122 ***
density:factor(refuge)1 1.07564 0.15807 6.805 1.89e-05 ***
density:factor(refuge)2 0.44875 0.07782 5.767 8.93e-05 ***
density:factor(refuge)3 -0.02728 0.12818 -0.213 0.835038
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.2901 on 12 degrees of freedom
Multiple R-squared: 0.985, Adjusted R-squared: 0.9775
F-statistic: 131.2 on 6 and 12 DF, p-value: 3.13e-10
Based on the labels of the coefficient vector of regression parameters I write a function that constructs the appropriate vector of regressors corresponding to the given values of density and refuge.
Our goal is to calculate the confidence level that allows individual overlapping confidence intervals to be used to graphically assess treatment differences separately at each value of goby density. To accomplish this we need the ci.func function that was first presented in lecture 5.
This function requires the variance-covariance matrix of the means. I illustrate how to create it for the smallest goby density when refuge = 1.
I build a data frame consisting of the three values of refuge density with goby density = 1.125.
I construct a vector of regressors for each of the three observations in the data frame and assemble them in a matrix.
I obtain the variance-covariance matrix of the means and use it as input to the ci.func function to obtain the difference-adjusted confidence levels needed to make pairwise comparisons between the three refuge category means.
I assemble the above steps in a function so that we can obtain the difference-adjusted confidence levels using any goby density value.
Finally I use this function on each of the different goby density values using sapply to apply the function separately to each density value.
The range of confidence levels reported is from 0.85 to 0.87 so there isn't much variability. Clearly we can get by with using just one value. I add 95% and 85% confidence intervals for the mean to each point on the regression line that corresponds to an observed goby density. To illustrate the steps I start by doing just refuge = 1.
Using the means and standard errors I plot the regression line, 95% confidence intervals of the mean at each goby density value for refuge = 1, and the 85% difference-adjusted intervals.
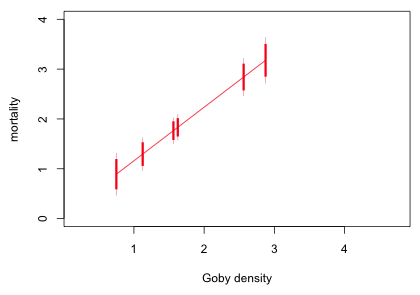 |
| Fig. 10 Preliminary graph showing confidence intervals for refuge = 1 |
Finally I assemble things as a function whose argument is the value of refuge. I include two color arguments to specify colors for the regression line and the difference-adjusted confidence levels.
Finally I draw the graph.
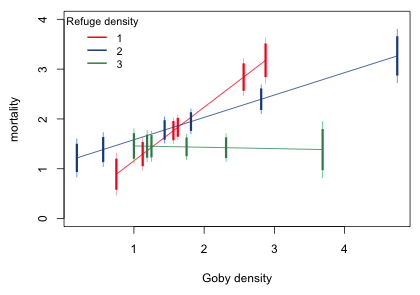 |
| Fig. 11 Graph to compare mean mortality across refuge level for specific values of goby density |
What we can see from Fig. 11 is that when goby density > 1.8 (approximately) refuge = 3 has a significantly lower mean mortality than do the other two values of refuge. The sparse data make it difficult to determine these change point precisely but probably when goby density > 2.8, refuge = 2 has a significantly lower mortality than does refuge = 1.
A compact collection of all the R code displayed in this document appears here.
| Jack Weiss Phone: (919) 962-5930 E-Mail: jack_weiss@unc.edu Address: Curriculum for the Environment and Ecology, Box 3275, University of North Carolina, Chapel Hill, 27599 Copyright © 2012 Last Revised--September 30, 2012 URL: https://sakai.unc.edu/access/content/group/3d1eb92e-7848-4f55-90c3-7c72a54e7e43/public/docs/lectures/lecture9.htm |