Historically a quantity called the deviance has played a large role in assessing the fit of generalized linear models. The deviance derives from the usual likelihood ratio statistic as we now illustrate. Suppose we have two nested models with estimated parameter sets ![]() and
and ![]() and corresponding likelihoods
and corresponding likelihoods ![]() and
and ![]() . If
. If ![]() is a special case of
is a special case of ![]() then we can compare the models using a likelihood ratio test.
then we can compare the models using a likelihood ratio test.
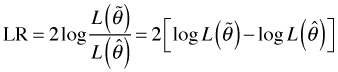
Recall that LR has an asymptotic chi-squared distribution with degrees of freedom equal to the difference in the number of parameters estimated in the two models. Alternatively, the degrees of freedom is the number of parameters that are fixed to have a specific value in one model but are freely estimated in the other model.
A saturated model is one in which one parameter is estimated for each observation. Suppose the model with likelihood ![]() is the saturated model and
is the saturated model and ![]() is the likelihood of some model of interest, call it model M. In the special case where model M is being compared to the saturated model, the likelihood ratio statistic for this comparison is called the deviance. Technically a distinction is made between the residual deviance, DM , and the scaled deviance, the residual deviance divided by the scale parameter.
is the likelihood of some model of interest, call it model M. In the special case where model M is being compared to the saturated model, the likelihood ratio statistic for this comparison is called the deviance. Technically a distinction is made between the residual deviance, DM , and the scaled deviance, the residual deviance divided by the scale parameter.
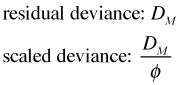
The scale parameter is a term recognizable in the log-likelihood when it is written in its exponential form. For the Poisson and binomial distributions, the two probability models for which the deviance can be legitimately used as a goodness of fit statistic, this distinction between residual deviance and scaled deviance is unimportant because the scale parameter φ = 1, so the two are the same.
In the saturated model n parameters are estimated, one for each of the n observations. In model M there are p < n parameters that are estimated. This means that n – p of the parameters that were estimated in the saturated model are set to zero in Model M. Because the scaled deviance is a likelihood ratio statistic it follows that
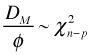
Thus the deviance has the form of a goodness of fit statistic. Large values of the deviance, relative to its degrees of freedom, indicate a significant lack of fit. As was noted above there are only two kinds of generalized linear models for which the deviance is an appropriate goodness of fit statistic: Poisson regression and logistic regression with grouped binary data (which we'll consider in lecture 14). The deviance statistic should not be used as a goodness of fit statistic for logistic regression with a binary response.
The mean of a chi-squared distribution is equal to its degrees of freedom, i.e., ![]() . Thus if a model provides a good fit to the data and the chi-squared distribution of the deviance holds, we expect the scaled deviance of the model to be close to its mean value n – p, i.e.,
. Thus if a model provides a good fit to the data and the chi-squared distribution of the deviance holds, we expect the scaled deviance of the model to be close to its mean value n – p, i.e., 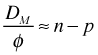 . For Poisson and binomial probability models, φ = 1. Thus for these models we expect
. For Poisson and binomial probability models, φ = 1. Thus for these models we expect ![]() or equivalently
or equivalently 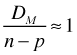 . So a quick test of the adequacy of a Poisson or binomial model is to divide the residual deviance by its degrees of freedom and see if the result is close to one. If the ratio is much larger than one we call the data overdispersed. If it is much smaller than one we call the data underdispersed. (Note: The use of these terms should be restricted to Poisson and binomial distributions.)
. So a quick test of the adequacy of a Poisson or binomial model is to divide the residual deviance by its degrees of freedom and see if the result is close to one. If the ratio is much larger than one we call the data overdispersed. If it is much smaller than one we call the data underdispersed. (Note: The use of these terms should be restricted to Poisson and binomial distributions.)
Unfortunately the accuracy of this test is somewhat dubious with small data sets. In order for the chi-squared distribution of the deviance to hold, the expected Poisson or binomial counts, i.e., the fitted values predicted by the model, should be large. A general rule is that they should exceed 5, although when there are many categories Agresti (2002) notes that an expected cell frequency as small as 1 is okay as long as no more than 20% of the expected counts are less than 5.
To illustrate the use of the deviance for checking model fit we revisit a Poisson regression model that was fit in lecture 11.
To fit the saturated model we create a variable that has a different value for each observation, convert it to a factor, and then use it as a predictor in a Poisson regression model.
So in the saturated model we have 280 parameters, one for each observation. To compare our model to the saturated model we can use a likelihood ratio test.
The degrees of freedom of the likelihood ratio statistic is just the difference in number of parameters fit in the two models.
This likelihood ratio statistic and its degrees of freedom are reported as part of the summary output from the model and labeled the residual deviance.
Deviance Residuals:
Min 1Q Median 3Q Max
-12.0267 -2.6401 -1.5599 0.1912 20.9158
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.0349466 0.0911488 -0.383 0.701
CORAL_COVE 0.0332184 0.0018308 18.144 <2e-16 ***
WSSTA 0.3056099 0.0162077 18.856 <2e-16 ***
I(WSSTA^2) -0.0270397 0.0007945 -34.032 <2e-16 ***
CORAL_COVE:WSSTA 0.0038035 0.0002341 16.246 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for poisson family taken to be 1)
Null deviance: 11827.0 on 279 degrees of freedom
Residual deviance: 4912.3 on 275 degrees of freedom
AIC: 5527.6
Number of Fisher Scoring iterations: 7
To carry out the deviance goodness of fit test we compare the deviance to the critical value of a chi-squared distribution with 275 degrees of freedom. The p-value of the residual deviance is
Because the p-value is very small, we should reject the null hypothesis. Before we conclude that we have a significant lack of fit, we should verify that our data meet the minimum sample size criteria. We calculate the expected counts and determine the fraction of them that are less than 5.
So 55% of the expected cell counts are less than five. This is far greater than 20% suggesting that we should not take the deviance test seriously here.
The likelihood ratio test is often written with a negative sign.
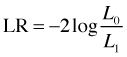
Written this way L1 is the larger of the two models (it has more estimated parameters) and L0 is the simpler model in which some of those parameters have been assigned specific values (such as zero). Written this way and letting LS denote the likelihood of the saturated model, the scaled deviance can be written as follows.
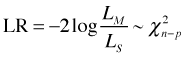
Now suppose we wish to compare two nested models with likelihoods L1 and L2 in which L2 is the "larger" (more estimated parameters) of the two likelihoods. The likelihood ratio statistic for comparing these two models, written in the alternate format given above, is the following.
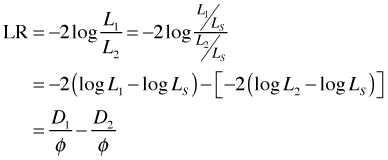
where D1 and D2 are the deviances of models 1 and 2, respectively. Thus to carry out a likelihood ratio test to compare two models we can equivalently just compute the difference of their scaled deviances.
Because of the parallel to analysis of variance (ANOVA) in ordinary linear models, carrying out an LR test using the scaled deviances of the models is often called analysis of deviance. This is what is reported by the anova function in R. The column labeled "Deviance" really represents the change in deviance between two adjacent models in the table. These are also likelihood ratio statistics.
Model: poisson, link: log
Response: PREV_1
Terms added sequentially (first to last)
Df Deviance Resid. Df Resid. Dev Pr(>Chi)
NULL 279 11827.0
CORAL_COVE 1 4617.5 278 7209.5 < 2.2e-16 ***
WSSTA 1 51.4 277 7158.0 7.371e-13 ***
I(WSSTA^2) 1 1967.7 276 5190.4 < 2.2e-16 ***
CORAL_COVE:WSSTA 1 278.1 275 4912.3 < 2.2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
In the ordinary normal-based multiple regression model, we write
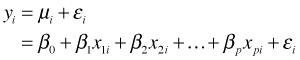
where εi is referred to as the error. When we obtain estimates for the parameters we write the fitted model as
![]()
so that ![]() is an estimate of the errors, εi. The ei are called residuals, or more properly, the raw residuals (also called the response residuals) of the model. For generalized linear models the raw residuals are seldom useful. Instead two other kinds of residuals, deviance residuals and Pearson residuals, are used.
is an estimate of the errors, εi. The ei are called residuals, or more properly, the raw residuals (also called the response residuals) of the model. For generalized linear models the raw residuals are seldom useful. Instead two other kinds of residuals, deviance residuals and Pearson residuals, are used.
The deviance is twice the difference in log-likelihoods between the current model and a saturated model.
![]()
Because the likelihood is a product of probabilities or densities, each log-likelihood is a sum of a probability or density for each observation. If we group the corresponding log-likelihood terms from both the model and the saturated model by observation, we get that observation's contribution to the deviance which we denote by ![]() . Because of the factor of two that multiplies the difference in logs, each of these contributions is squared,
. Because of the factor of two that multiplies the difference in logs, each of these contributions is squared, ![]() . The deviance residual is defined as follows.
. The deviance residual is defined as follows.
![]()
Here sign denotes the sign function which is defined as follows.
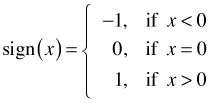
Because under ideal conditions the deviance can be used as a measure of lack of fit, the deviance residual measures the ith observation's contribution to model lack of fit.
The Pearson residual is a standardized raw residual. The raw residual is divided by an estimate of the standard error of the prediction.
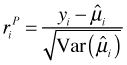
This estimate will vary depending on the probability distribution chosen for the response. The Pearson residual is that observation's contribution to the Pearson goodness of fit statistic.
A binomial random variable is discrete. It records the number of successes out of n trials. The classic illustration of the binomial distribution is an experiment in which we record the number of heads obtained when a coin is flipped n times. A binomial random variable is bounded on both sides—below by 0, above by n. This distinguishes it from a Poisson random variable which is bounded below by zero but is unbounded above.
An experiment is called a binomial experiment if four assumptions are met.
The binomial is a two-parameter distribution with parameters n and p. We write![]() . The mean and variance are
. The mean and variance are
Observe from this last expression that the variance is a function of the mean, i.e., ![]() . If you plot the variance of the binomial distribution against the mean you obtain a parabola opening downward with a maximum at
. If you plot the variance of the binomial distribution against the mean you obtain a parabola opening downward with a maximum at ![]() corresponding to p = 0.5. When n = 1 the binomial distribution is sometimes called the Bernoulli distribution and the data are referred to as binary data. To contrast this situation from the binomial distribution proper, we will refer to data arising from a binomial distribution with n > 1 as grouped binary data.
corresponding to p = 0.5. When n = 1 the binomial distribution is sometimes called the Bernoulli distribution and the data are referred to as binary data. To contrast this situation from the binomial distribution proper, we will refer to data arising from a binomial distribution with n > 1 as grouped binary data.
Examples of binomial data
R binomial functions are denoted: dbinom, pbinom, qbinom, rbinom. In R the parameters n and p are specified with the argument names size and prob respectively. There is no special function for binary data in R. Just use the binomial function and set size = 1 (n = 1).
Logistic regression is the preferred method of analysis for situations in which the response has a binomial distribution. Logistic regression is a generalized linear model in which
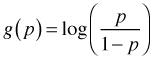 , also called the logit.
, also called the logit.When the binomial distribution is put in its standard exponential form the parameter p only occurs as part of the logit function 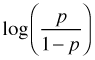 . For this reason the logit function is called the canonical link in logistic regression. The logit turns out to be an excellent choice as a link function. One of the problems with using the identity link for proportions is that although proportions are required to lie in the interval [0, 1], an identity link can yield predictions outside this interval. The logit function restricts p to the unit interval as we now demonstrate. Setting the logit link equal to the linear predictor and solving for p yields the following.
. For this reason the logit function is called the canonical link in logistic regression. The logit turns out to be an excellent choice as a link function. One of the problems with using the identity link for proportions is that although proportions are required to lie in the interval [0, 1], an identity link can yield predictions outside this interval. The logit function restricts p to the unit interval as we now demonstrate. Setting the logit link equal to the linear predictor and solving for p yields the following.
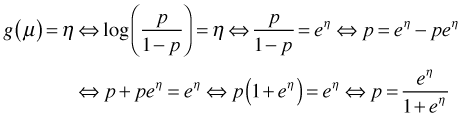
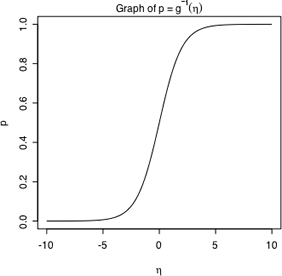
Fig. 1 Inverse of the logit link
The expression 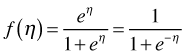 that inverts the logit link is called the logistic function and is shown in Fig. 1. Observe the following.
that inverts the logit link is called the logistic function and is shown in Fig. 1. Observe the following.
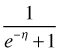 is also always positive.
is also always positive. 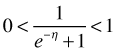 .
.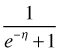 approaches 0 as
approaches 0 as 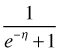 approaches 1 as
approaches 1 as Other suitable link functions for binomial data are the probit, ![]() , and the complementary log-log link,
, and the complementary log-log link, ![]() , but the logit is certainly the most popular choice. The reason the logit is the preferred link function for binomial data is that it is interpretable. We examine this interpretation next.
, but the logit is certainly the most popular choice. The reason the logit is the preferred link function for binomial data is that it is interpretable. We examine this interpretation next.
As a background for understanding how to interpret regression coefficients in models with a logit link, we review the interpretation of regression coefficients in other kinds of regression models.
When ![]() is the identity function, as is typically the case in ordinary linear regression, then
is the identity function, as is typically the case in ordinary linear regression, then
![]() .
.
If we increase x1 by one unit, then we have
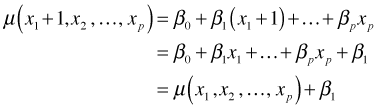
So β1 is interpreted as the amount that the mean is increased when x1 is increased by one unit.
When ![]() is the log function, as in Poisson or negative binomial regression, then
is the log function, as in Poisson or negative binomial regression, then
![]()
If we increase x1 by one unit, then we have
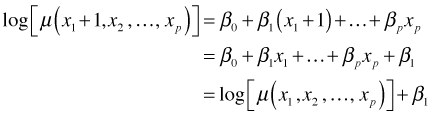
So β1 is the amount that the log mean is increased when x1 is increased by one unit. Converting to the original scale of the mean we have the following.
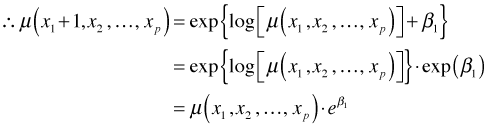
So we can interpret β1 in terms of ![]() , which is the amount the mean is multiplied by when x1 is increased by one unit.
, which is the amount the mean is multiplied by when x1 is increased by one unit.
The argument just given in which we invert the identity and log links does not work for the logit link. The regression coefficients do not have simple interpretations on the scale of p, the probability of success. On the other hand the regression coefficients in logistic regression do have a simple interpretation in terms of the odds. Probability theory had its origin in the study of gambling. Gamblers typically speak in terms of the "odds for" or "odds against" events occurring, rather than in terms of probabilities, but the two notions are equivalent. Any statement made using odds can also be expressed in terms of probability.
As an example, suppose it is stated that the odds in favor of an event Y = 1 occurring are 3 to 1. This can be interpreted as follows.
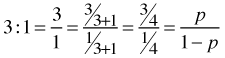
Thus if the odds for an event are 3:1, then the event has a probability ![]() of occurring.
of occurring.
Notice that it is odds, rather than probability, that is the argument of the logarithm function in the logit link. Thus the logit function is also called the log odds function. When the logit is viewed as a log odds, this immediately provides us with an interpretation for the regression parameter β1 in the logistic regression model. Switching to the log odds notation we have the following.
![]()
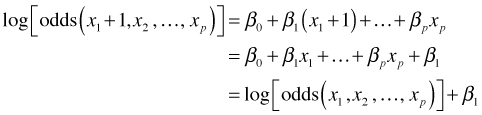
Solving for β1 yields the following.
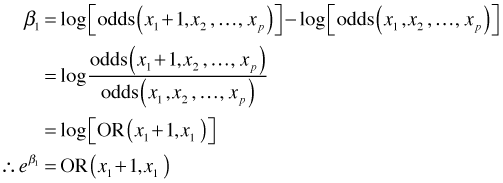
where the notation OR denotes the odds ratio, the ratio of two odds. The specific odds ratio shown here measures the effect of increasing x1 by 1 on the odds that Y = 1.
Just as is the case with ordinary regression, Poisson regression, and negative binomial regression, we can use the sign of the regression coefficient in logistic regression to infer the effect a predictor has on the response. If β1 is the regression coefficient of x1in the logistic regression model we have the following.
| Jack Weiss Phone: (919) 962-5930 E-Mail: jack_weiss@unc.edu Address: Curriculum for the Environment and Ecology, Box 3275, University of North Carolina, Chapel Hill, 27599 Copyright © 2012 Last Revised--February 13, 2012 URL: https://sakai.unc.edu/access/content/group/2842013b-58f5-4453-aa8d-3e01bacbfc3d/public/Ecol562_Spring2012/docs/lectures/lecture13.htm |