Observational heterogeneity occurs when some observations resemble others purely because of the way the data were collected.

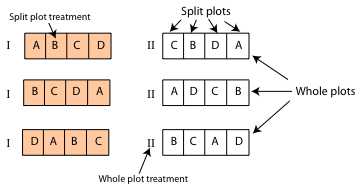
Fig. 1 Split plot design
Any time we have a lack of equivalence between the observational or experimental units used in a study, we have observational heterogeneity. Last week we discussed one specific approach for dealing with such heterogeneity—generalized least squares (GLS). We focused on the specific case of repeated measures data in which there is a long time series of data, but generalized least squares can be used any time data are organized in a hierarchical fashion. Unfortunately generalized least squares has some limitations.
Today we discuss a second approach for dealing with observational heterogeneity in regression models—introducing random effects to produce what's called a mixed effects model. Mixed effects models are an omnibus way to account for observational heterogeneity. To set up a mixed model we just need to know how the data are structured, i.e., be able to identify the different sized units in the analysis. We don't actually have to understand the precise nature of the relationships (correlations) of the observations that make up the different sized units. Thus mixed effects models are a convenient way of addressing data structure especially in situations where the structure is a nuisance and is of little interest to us by itself. On the other hand if we fit a mixed effects model to temporal data it may still be necessary to account for lingering residual temporal correlation.
As an illustration of these basic ideas we return to the coral core data set we analyzed using generalized least squares last time. The basic goal was to model how coral extension rates (the widths of annual rings in coral cores) vary over time. The question of interest is whether extension rates have shown a linear trend over time that depends upon the location of the coral colony in the reef complex (nearshore, forereef, and backreef locations). We assume that extension rates are normally distributed with a mean that may be changing with time. Thus our basic assumption is ![]() where i denotes a coral core and j denotes an individual observation (annual ring) from that coral core.
where i denotes a coral core and j denotes an individual observation (annual ring) from that coral core.
In the common pooling model we ignore the structure of the data entirely. We treat all of the observations as coming from a single population from which we've drawn a single random sample. For the coral core data set we would start by assuming that the mean ![]() is a linear function of calendar year.
is a linear function of calendar year.
![]()
where again i = core and j = individual observation from that core. The problem with the common pooling model is that it is almost certainly false. The errors are not independent as we saw last time. By ignoring data structure and treating the individual rings as being a random sample from the population of coral core annual rings we are guilty of pseudo-replication, claiming that we have an effective sample size that is much larger than the one we really have.
In the fixed effects approach to structured data, we include the structural variable as a predictor in the model. In the current example that translates into specifying dummy variables for individual cores and including them as additive terms and as interaction terms with year. This yields a separate intercept and slope for each core.
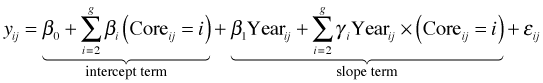
where g is the number of cores. As before, ![]() . The parameters β0 and β1 are the intercept and slope for core 1. βi and γi are the deviations that the intercept and slope of core i exhibit from the intercept and slope of core 1. This single model is comparable to fitting separate regression models to each core except that when we do it using dummy variables in a single regression model we use all of the data to estimate the residual variance σ2.
. The parameters β0 and β1 are the intercept and slope for core 1. βi and γi are the deviations that the intercept and slope of core i exhibit from the intercept and slope of core 1. This single model is comparable to fitting separate regression models to each core except that when we do it using dummy variables in a single regression model we use all of the data to estimate the residual variance σ2.
Although this approach gets the structure of the data set correct, something that was ignored in the common pooling model, it has other problems.
Even though in lecture 17 we were able to simplify the separate slopes and intercepts model so that we needed to estimate only three slopes, one for each reef type, we were still left with estimating separate intercepts for each core. If there had been more cores used in the analysis it is unlikely we would have been able to make even this simplification. Fitting separate models to individual natural groups will nearly always provide a significantly better fit to data than will any simpler model that we can construct. The basic problem with the fixed effects approach is that it typically leads to overfitting the data.
The random effects analog of the separate slopes and intercepts regression model is the random slopes and intercepts model.
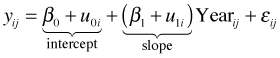
with ![]() . In this model β0 and β1 represent the population-average coefficients while
. In this model β0 and β1 represent the population-average coefficients while ![]() and
and ![]() are the deviations from this population average for coral core i. β0 and β1 can also be interpreted as the regression coefficients for a typical core, i.e., one corresponding to the middle of the distribution of random effects. The intercept for core i is
are the deviations from this population average for coral core i. β0 and β1 can also be interpreted as the regression coefficients for a typical core, i.e., one corresponding to the middle of the distribution of random effects. The intercept for core i is ![]() and the slope is
and the slope is ![]() . Thus the random slopes and intercepts formulation of this model is also the following.
. Thus the random slopes and intercepts formulation of this model is also the following.
![]()
What makes this model different from the fixed effects model is that ![]() and
and ![]() are not directly estimated but instead are assumed to be drawn from a multivariate normal distribution.
are not directly estimated but instead are assumed to be drawn from a multivariate normal distribution.
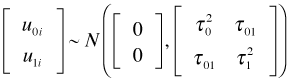
The diagonal entries of the multivariate normal covariance matrix are the individual variances of the intercept and slope random effects and the off-diagonal entry is their covariance. Because the correlation coefficient is defined by 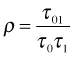 , an equivalent way of writing this distribution (and the one used by R) is the following.
, an equivalent way of writing this distribution (and the one used by R) is the following.
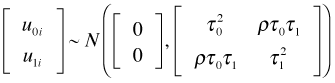
Rather than estimate the individual ![]() and
and ![]() we instead estimate the parameters of the covariance matrix of the multivariate normal distribution: ρ, τ0, and τ1.
we instead estimate the parameters of the covariance matrix of the multivariate normal distribution: ρ, τ0, and τ1.
| Jack Weiss Phone: (919) 962-5930 E-Mail: jack_weiss@unc.edu Address: Curriculum for the Environment and Ecology, Box 3275, University of North Carolina, Chapel Hill, 27599 Copyright © 2012 Last Revised--February 20, 2012 URL: https://sakai.unc.edu/access/content/group/2842013b-58f5-4453-aa8d-3e01bacbfc3d/public/Ecol562_Spring2012/docs/lectures/lecture18.htm |